Please note, this documentation is a work in progress. Like much else of the system. That means you are free to edit it as well as include new sections that you think are pertinent. It also isn't likely to be completely up to date
Also all programs written by me that form part of this system are licensed under the general public license (GPL). Basically that means that you are free to redistribute the programs, as long as you make the source code available and you redistribute under the GPL. For more information see the license and the Free Software Foundation for more information. For contact details see the bottokm of this page.
The eXintegrator system aims to provide rapid and easy access to microarray expression data from a large number of sources and to integrate this data with externally incoporated biological data. This website is divided into the following sections:
I'm not particularly happy with the name given to the system at the moment so if anyone has some good suggestion as to a better name, then please let me know. It's not so easy to find a reasonable name for anything these days, Google always seems to find some prior Trademarked use of the name.
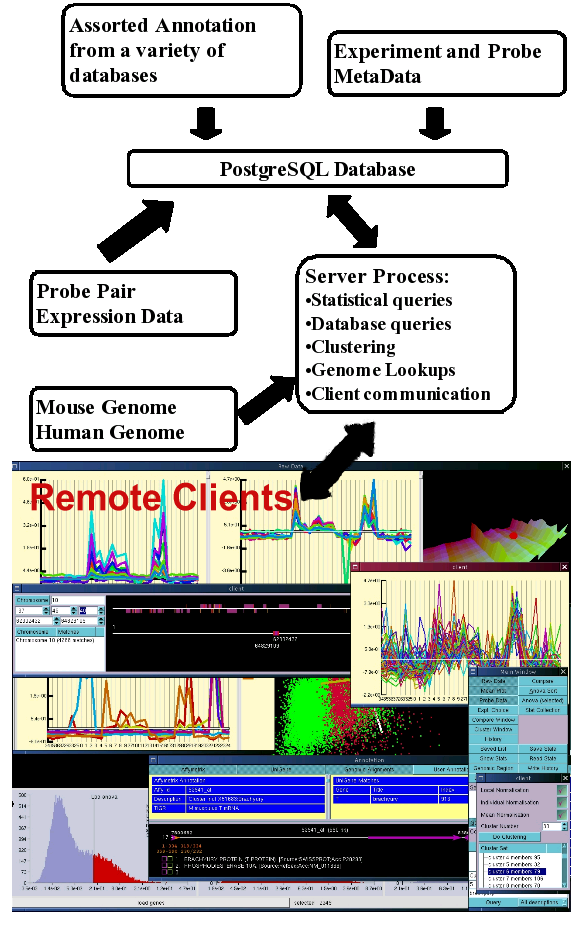
In order to allow many users to access microarray data quickly the system was designed as a client server system where the server maintains all the expression data along with associated information in memory. This server process communicates with an underlying database which it uses to make queries on associated data as well as to store user selections and annotation. The server process communicates with client applications which are used mainly as viewers and query (both statistical and database) input mechanisms. This means that users do not have to load large amounts of data into memory in order to perform analyses, and since the expression data is kept in memory, queries on the data can be performed very rapidly.
The underlying database also includes probe set annotation from a number of sources. This probe set annotation also includes genomic loci to which the probe sets map. This allows the probe sets not only to be linked to a some set of annotation, but also to a given loci, which in turn can be linked to additional information linked to that loci. The system presents expression and annotation data simultaneously to quickly allow users to select interesting differences in expression. In addition since the system includes information from an extended data set any differences in expression observed are put into a wider context of specifity and variability of expression of genes viewed.
The eXintegrator system. Copyright (C) 2004 Martin Jakt & Okada Mitsuhiro
Martin Jakt mjakt@cdb.riken.jp
Okada Mitsuhiro mitsu@cdb.riken.jp
Stem Cell Biology Group
The Foundation for Biomedical Research and Innovation
&
RIKEN Center for Developmental Biology
2-2-3 Minatojima-minamimachi
Chuo-ku, Kobe, Hyogo 650-0047
Japan