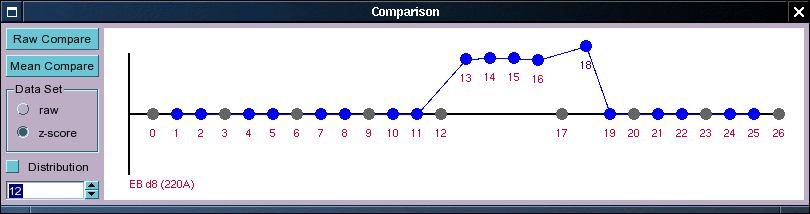
One of the most common requests by researchers is to find a set of probe sets (or rather genes) which have some specific expression pattern. Our approach to this is to allow the researcher to specify the pattern desired through a graphical input and then to merely sort the probe sets by their euclidean distance to the specified expression profile. Rather than choosing some threshold distance we urge the user merely to use the system to go through the probe sets until there clearly is no tendency of the probe sets to show a similar expression profile. This is partly because the euclidean distance may not be a particularly good statistic for choosing a 'good' cut-off, but also because if the pattern specified is complex then there are many different ways in which the pattern can deviate from the specified one. As it is difficult for most researches to specify what kinds of deviations are ok, and which ones are less so we generally think that it is better to select interesting probe sets by eye and gut feeling rather than by strict statistical parameters. This it should be noted is not the best way to select probe sets if the question asked is a simple binary one (i.e. higher in sample a than in sample b), but because going through the samples manually will also display the expression pattern of the probe set in an extended data set as well as the probe set annotation, we believe that this approach still has its uses in these kind of cases (there are ways to combine these approaches with external programs, but this will not be discussed here).