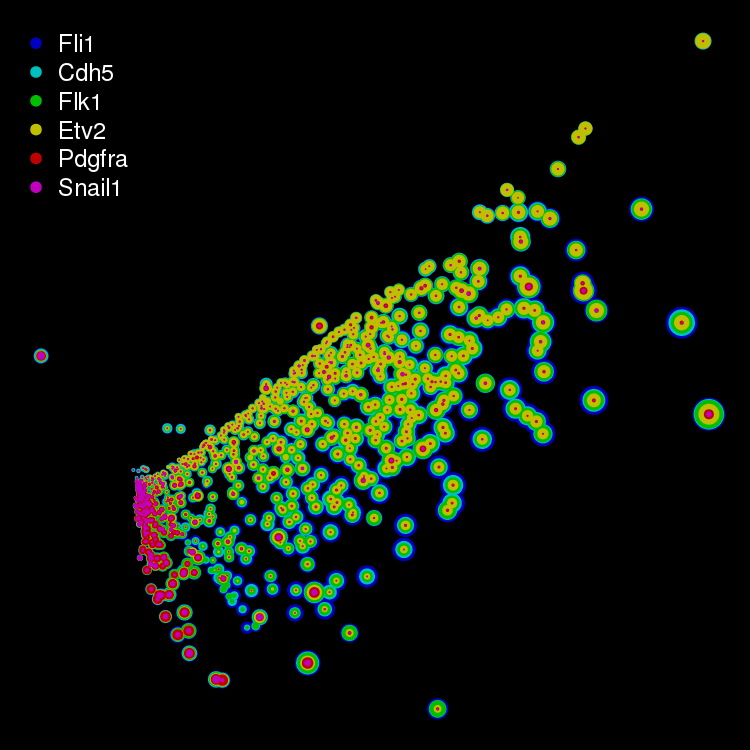
Etv2 and its direct targets
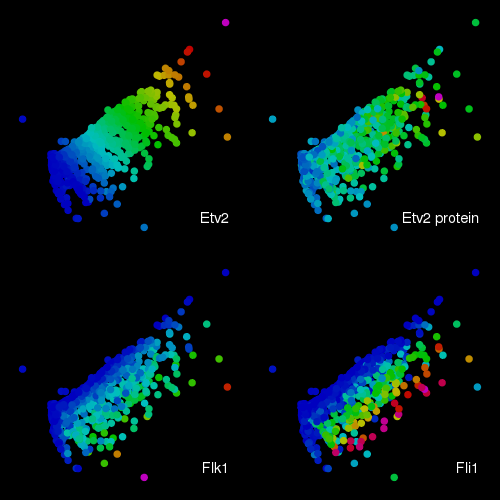
Transcripts from a dox inducible EGFP-Etv2 transcription factor and the Flk1,
Fli1, Cdh5, Snail1 and Pdgfra genes were enumerated by combinatorial FISH. In
addition, the amount of EGFP-Etv2 fusion protein was estimated from direct EGFP
fluorescence. The transcript densities were used to determine the global set
of relationships between the cells and these were reduced from 6 to 2 dimensions
using the SOD algorithm. The colouring in the individual plots indicate the
relative levels of either transcriptional densities or the estimated EGFP-Etv2
protein amounts.
Click image for the scalable svg file.